# from app import app, db#, cache
from matplotlib.mlab import griddata
import matplotlib.pyplot as plt
import time, os
from collections import defaultdict
import numpy as np
from numpy import linspace
from numpy import meshgrid
from numpy.random import uniform, seed
from mpl_toolkits.basemap import Basemap, shiftgrid
from mpl_toolkits.axes_grid1 import make_axes_locatable
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
title_sz = 27
axis_sz = 22
tick_sz = 21
from matplotlib.ticker import MaxNLocator
from matplotlib import cm, rcParams
rcParams.update({'font.size': tick_sz-4}) # Increase font-size
# from matplotlib.backends.backend_pdf import PdfPages
import matplotlib.cm as cm
from matplotlib import gridspec
import pylab as pl
from itertools import islice
import sys, pickle, time, copy, re
from chem_ocean import Build_Map as bm
import sqlalchemy
import pandas as pd
Visualizing cluster centers given added geo features#
from sklearn.cluster import MiniBatchKMeans as mkm
from sklearn import preprocessing
from sklearn.preprocessing import StandardScaler
def connect(user, password, db, host='localhost', port=5432):
'''Returns a connection and a metadata object'''
url = 'postgresql://{}:{}@{}:{}/{}'
url = url.format(user, password, host, port, db)
# The return value of create_engine() is our connection object
con = sqlalchemy.create_engine(url, client_encoding='utf8')
# We then bind the connection to MetaData()
meta = sqlalchemy.MetaData(bind=con, reflect=True)
return con, meta, url
conn, meta, url = connect('jlanders', '', 'odv_app_dev', host='localhost', port=5432)
/usr/local/lib/python3.6/site-packages/psycopg2/__init__.py:144: UserWarning: The psycopg2 wheel package will be renamed from release 2.8; in order to keep installing from binary please use "pip install psycopg2-binary" instead. For details see: <http://initd.org/psycopg/docs/install.html#binary-install-from-pypi>.
""")
/usr/local/lib/python3.6/site-packages/ipykernel_launcher.py:10: SADeprecationWarning: reflect=True is deprecate; please use the reflect() method.
# Remove the CWD from sys.path while we load stuff.
minLat = -55
maxLat = 60
minLon = -180
maxLon = 180
# Helper function to query DataFrame for slice of interest
def dfQueryFunc(minLat, maxLat, minLon, maxLon, _var_names,**kwargs):
basic = ['station', 'longitude', 'latitude']
if 'Depth' in kwargs:
sum_names = basic +_var_names
else:
sum_names = basic + ['depth'] + _var_names
print(sum_names)
cols = ', '.join(sum_names)
if 'Latitude' in kwargs:
lat_target = kwargs['Latitude']
minLat = lat_target-1.5
maxLat = lat_target+1.5
(lonMin, lonMax) = kwargs['lonLimits']
# _var_df = _df3_sub[sum_names].query('Latitude >= '+str(lat_target-3)+ 'and Latitude <= '+str(lat_target+3)+ ' and Longitude >= '+str(lonMin)+ 'and Longitude <= '+str(lonMax))
if 'Longitude' in kwargs:
lon_target = kwargs['Longitude']
minLon = lon_target-1.5
maxLon = lon_target+1.5
(latMin, latMax) = kwargs['latLimits']
# _var_df = _df3_sub[sum_names].query('Longitude >= '+str(lon_target-3)+ 'and Longitude <= '+str(lon_target+3) +' and Latitude >= '+str(latMin)+ 'and Latitude <= '+str(latMax))
query = "select "+ cols+ " from woa13 where latitude> {} AND latitude< {} AND longitude>{} and longitude<{};".format(str(minLat), str(maxLat), str(minLon), str(maxLon))
if 'Depth' in kwargs:
depth_target = kwargs['Depth']
query = "select "+ cols+ " from woa13 where latitude> {} AND latitude< {} AND longitude>{} and longitude<{} and depth={};".format(str(minLat), str(maxLat), str(minLon), str(maxLon), str(depth_target))
print(query)
return query, sum_names#_var_df
tracers = ['salinity', 'oxygen', 'nitrate', 'temperature']
query, sum_names = dfQueryFunc(minLat, maxLat, minLon, maxLon, tracers, Depth=1500)
['station', 'longitude', 'latitude', 'salinity', 'oxygen', 'nitrate', 'temperature']
select station, longitude, latitude, salinity, oxygen, nitrate, temperature from woa13 where latitude> -55 AND latitude< 60 AND longitude>-180 and longitude<180 and depth=1500;
result = conn.execute(query)
data_array = []
for row in result:
data_array.append(list(row))
df = pd.DataFrame(data_array)
df.columns = sum_names
df.dropna()
lat_scale = StandardScaler()
lat_scale.fit(df[['latitude']].as_matrix())
df['s_latitude'] = lat_scale.transform(df[['latitude']])
lon_scale = StandardScaler()
lon_scale.fit(df[['longitude']].as_matrix())
df['s_longitude'] = lon_scale.transform(df[['longitude']])
for tracer in tracers:
df[tracer].round(decimals = 3)
scale = StandardScaler()
df[[tracer]] = scale.fit_transform(df[[tracer]].as_matrix())
df.describe()
| longitude | latitude | salinity | oxygen | nitrate | temperature | s_latitude | s_longitude | |
|---|---|---|---|---|---|---|---|---|
| count | 26361.000000 | 26361.000000 | 2.636100e+04 | 2.636100e+04 | 2.636100e+04 | 2.636100e+04 | 2.636100e+04 | 2.636100e+04 |
| mean | -14.016217 | -7.010944 | -2.577911e-15 | -1.983838e-16 | 3.795168e-16 | -4.808650e-16 | -1.725076e-17 | -2.156345e-18 |
| std | 111.438920 | 30.964058 | 1.000019e+00 | 1.000019e+00 | 1.000019e+00 | 1.000019e+00 | 1.000019e+00 | 1.000019e+00 |
| min | -179.500000 | -54.500000 | -2.444244e+01 | -2.257720e+00 | -4.594790e+00 | -2.760163e+00 | -1.533712e+00 | -1.485001e+00 |
| 25% | -118.500000 | -34.500000 | -2.362516e-01 | -8.329838e-01 | -3.065452e-01 | -4.126793e-01 | -8.877899e-01 | -9.376057e-01 |
| 50% | -25.500000 | -9.500000 | -1.524248e-01 | 1.408474e-01 | 1.524210e-01 | -2.182675e-01 | -8.038685e-02 | -1.030520e-01 |
| 75% | 83.500000 | 17.500000 | 9.648754e-02 | 7.312365e-01 | 7.339001e-01 | 2.275435e-01 | 7.916084e-01 | 8.750809e-01 |
| max | 179.500000 | 59.500000 | 8.061613e+00 | 2.396203e+00 | 1.977760e+00 | 9.611661e+00 | 2.148046e+00 | 1.736556e+00 |
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(17, 17), facecolor='w')
_basemap, fig, ax = bm.build_map('y', 'merc', minLat, maxLat, minLon, maxLon, 'c', fig, ax, 111, 'lb')
colors = ['bo', 'ro', 'go', 'co']
for ik, tracer in enumerate(tracers):
data = df[['s_longitude', 's_latitude', tracer]].values
kmeans = km(n_clusters=8, random_state=0).fit(data)
centers = kmeans.cluster_centers_
for center in centers:
x,y = _basemap(lon_scale.inverse_transform([center[0]]), lat_scale.inverse_transform([center[1]]))
ax.plot(x, y, colors[ik], markersize=10, alpha = .8, label = tracer)
from collections import OrderedDict
import matplotlib.pyplot as plt
handles, labels = plt.gca().get_legend_handles_labels()
by_label = OrderedDict(zip(labels, handles))
plt.legend(by_label.values(), by_label.keys(), loc = 3)
_x = df['longitude'].as_matrix()
_y = df['latitude'].as_matrix()
_feat_data= df['oxygen'].as_matrix()
x_coord,y_coord = _basemap(_x, _y)
cbar_pad = .22
# plt.tight_layout(pad=3, w_pad=4., h_pad=3.0)
ylabpad = 35
xlabpad = 25
# plt.tight_layout(pad=0.4, w_pad=1., h_pad=1.0)
# define grid.
xi = np.linspace(min(x_coord), max(x_coord),300)
yi = np.linspace(min(y_coord), max(y_coord),300)
# grid the data.
zi = griddata(x_coord,y_coord,_feat_data,xi,yi,interp='linear')
# contour the gridded data, plotting dots at the nonuniform data points.
CS = plt.contour(xi,yi,zi,10,linewidths=0.5,colors='k')
CS = plt.contourf(xi,yi,zi,40,cmap=plt.cm.brg,
vmax=2*abs(zi).max(), vmin=abs(zi).min())
# plt.legend(loc=3)
0.34735774993896484
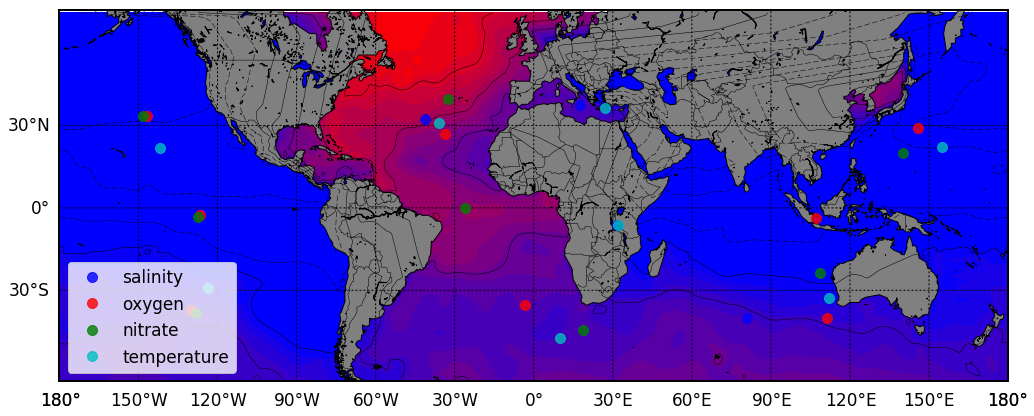
# Returns data for specific variables for relevant spatial constraints
# uses: dfQueryFunc
# slice type: plan, NSsection, EWsection
# kwargs: plan: depth; NSsection: lonTraj, latLImits; EWsection: latTrag, lonLimits
def amassData(minLat, maxLat, minLon, maxLon, _in_var_names, _sliceType, **kwargs):
DB_NAME = os.environ['DB_NAME']
DB_USER = os.environ['DB_USER']
DB_PASS = os.environ['DB_PASS']
DB_SERVICE = os.environ['DB_SERVICE']
DB_PORT = os.environ['DB_PORT']
SQLALCHEMY_DATABASE_URI = 'postgresql://{0}:{1}@{2}:{3}/{4}'.format(DB_USER, DB_PASS, DB_SERVICE, DB_PORT, DB_NAME)
# conn, meta, url = connect('jlanders', '', 'odv_app_dev2', host='localhost')
conn = sqlalchemy.create_engine(SQLALCHEMY_DATABASE_URI, client_encoding='utf8')
print(_sliceType)
# Depth slice
if _sliceType == 'plan':
if 'depth' in kwargs:
_depth = kwargs['depth']
else:
_depth = 1500
query, cols = dfQueryFunc(minLat, maxLat, minLon, maxLon, _in_var_names, Depth = _depth)
# psql
t0 = time.time()
result = conn.execute(query)
cluster_d = defaultdict(list)
for row in result:
for ik, col in enumerate(cols):
cluster_d[col].append(row[ik])
_x = np.asarray(cluster_d['longitude'])
_y = np.asarray(cluster_d['latitude'])
t1 = time.time()
print('psql', t1-t0)
_xLab = 'Longitude (deg)'
_yLab = 'Latitude (deg)'
_basemap = True
_latLon_params = None
# North-South (longitudinal) slice
if _sliceType == 'NSsection':
if 'lonTraj' in kwargs:
_lonTraj = kwargs['lonTraj']
print('_lonTraj', _lonTraj)
else:
_lonTraj = -15
if 'latLimits' in kwargs:
_latLims = kwargs['latLimits']
else:
_latLims = [-55, -35]
query, cols = dfQueryFunc(minLat, maxLat, minLon, maxLon, _in_var_names, Longitude = _lonTraj, latLimits = _latLims)
# psql
t0 = time.time()
result = conn.execute(query)
cluster_d = defaultdict(list)
for row in result:
for ik, col in enumerate(cols):
cluster_d[col].append(row[ik])
_x = np.asarray(cluster_d['latitude'])
_y = np.asarray(cluster_d['depth'])
t1 = time.time()
print('psql', t1-t0)
_yLab = 'Depth (m)'
_xLab = 'Latitude (deg) along '+ r'%s $^\circ$' % abs(_lonTraj)+ ['W' if lon<0 else 'E' for lon in [_lonTraj]][0]
_basemap = False
_latLon_params = ((_lonTraj, _lonTraj), _latLims)
# East-West (latitudeinal) slice
if _sliceType == 'EWsection':
if 'latTraj' in kwargs:
_latTraj = kwargs['latTraj']
else:
_latTraj = -45
if 'lonLimits' in kwargs:
_lonLims = kwargs['lonLimits']
else:
_lonLims = [-10, 10]
query, cols = dfQueryFunc(minLat, maxLat, minLon, maxLon, _in_var_names, Latitude = _latTraj, lonLimits = _lonLims)
# psql
t0 = time.time()
result = conn.execute(query)
cluster_d = defaultdict(list)
for row in result:
for ik, col in enumerate(cols):
cluster_d[col].append(row[ik])
_x = np.asarray(cluster_d['longitude'])
_y = np.asarray(cluster_d['depth'])
t1 = time.time()
print('psql', t1-t0)
_yLab = 'Depth (m))'
_xLab = 'Longitude (deg) along '+ r'%s $^\circ$' % abs(_latTraj)+ ['S' if lat<0 else 'N' for lat in [_latTraj]][0]
_basemap = False
_latLon_params = (_lonLims, [_latTraj, _latTraj])
# _feat_data = cluster_df[_in_var_names].as_matrix()
_feat_data = np.zeros((len(cluster_d['station']), len(_in_var_names)))
for ik, name in enumerate(_in_var_names):
_feat_data[:, ik] = np.asarray(cluster_d[name])
mask = np.all(np.isnan(_feat_data), axis=1)
_x = _x[~mask]
_y = _y[~mask]
_feat_data = _feat_data[~mask]
if len(_in_var_names) == 1:
_feat_data = np.ravel(_feat_data)
return _x, _y, _feat_data, _basemap, _xLab, _yLab, _latLon_params
def make_section(ax, _colors, _x, _y, _latLon_params, model_raw):
if model_raw == 'model':
for im in range(len(_x)):
ax.plot(_x[im], _y[im], c = _colors[im], marker = 'o', markersize=15, alpha = .1)
ax.invert_yaxis()
axin = inset_axes(ax, width="35%", height="35%", loc=4)
inmap = Basemap(projection='ortho', lon_0=np.mean(_latLon_params[0]), lat_0=0,
ax=axin, anchor='NE')
inmap.fillcontinents()
inmap.drawcoastlines(color='k')
inmap.plot(_latLon_params[0], _latLon_params[1], '-k', linewidth=2 , latlon=True)
return ax
'''
Plots raw data either in section or plan.
If there is a basemap, x=lon, y=lat and data is from a depth slice (plan).
If not, x=lat or lon, y=depth.
Both plot types are colored by contour for a specific chemical constituent.
'''
# def plotRaw(_x, _y, _feat_data, _basemap, _xLab, _yLab, _latLon_params, _in_var_names):
def getRaw(minLat, maxLat, minLon, maxLon, _in_var_names, _sliceType, **kwargs):
units_dict = {'temperature': '$^\circ$C', 'oxygen': 'ml/l', 'aou': 'ml/l', 'longitude': '$^\circ$E', 'salinity': '(psu)', 'nitrate': '$\mu$mol/l', 'depth': 'm', 'phosphate': '$\mu$mol/l', 'latitude': '$^\circ$N', 'oxygen_saturation': '%'}
if _sliceType == 'NSsection':
kwargs['lonTraj'] = minLon
kwargs['latLimits'] = (minLat, maxLat)
if _sliceType == 'EWsection':
kwargs['latTraj'] = minLat
kwargs['lonLimits'] = (minLon, maxLon)
_x, _y, _feat_data, _basemap, xLab, yLab, _latLon_params = amassData(minLat, maxLat, minLon, maxLon, _in_var_names, _sliceType, **kwargs)
return _x, _y, _feat_data, _basemap, xLab, yLab, _latLon_params
def plotRaw(minLat, maxLat, minLon, maxLon, _in_var_names, _sliceType, **kwargs):
units_dict = {'temperature': '$^\circ$C', 'oxygen': 'ml/l', 'aou': 'ml/l', 'longitude': '$^\circ$E', 'salinity': '(psu)', 'nitrate': '$\mu$mol/l', 'depth': 'm', 'phosphate': '$\mu$mol/l', 'latitude': '$^\circ$N', 'oxygen_saturation': '%'}
if _sliceType == 'NSsection':
kwargs['lonTraj'] = minLon
kwargs['latLimits'] = (minLat, maxLat)
if _sliceType == 'EWsection':
kwargs['latTraj'] = minLat
kwargs['lonLimits'] = (minLon, maxLon)
_x, _y, _feat_data, _basemap, xLab, yLab, _latLon_params = amassData(minLat, maxLat, minLon, maxLon, _in_var_names, _sliceType, **kwargs)
if len(_feat_data.shape)>1:
print( 'Please limit feature data to one variable')
else:
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(17, 17), facecolor='w')
if _basemap:
_basemap, fig, ax = bm.build_map('y', 'merc', minLat, maxLat, minLon, maxLon, 'c', fig, ax, 111, 'lb')
x_coord,y_coord = _basemap(_x, _y)
cbar_pad = .22
plt.tight_layout(pad=3, w_pad=4., h_pad=3.0)
ylabpad = 35
xlabpad = 25
plt.tight_layout(pad=0.4, w_pad=1., h_pad=1.0)
else:
x_coord = _x
y_coord = _y
ylabpad = 0
xlabpad = 0
# define grid.
xi = np.linspace(min(x_coord), max(x_coord),300)
yi = np.linspace(min(y_coord), max(y_coord),300)
# grid the data.
zi = griddata(x_coord,y_coord,_feat_data,xi,yi,interp='linear')
# contour the gridded data, plotting dots at the nonuniform data points.
CS = plt.contour(xi,yi,zi,10,linewidths=0.5,colors='k')
CS = plt.contourf(xi,yi,zi,15,cmap=plt.cm.rainbow,
vmax=abs(zi).max(), vmin=abs(zi).min())
# label axes
ax.set_ylabel(yLab, fontsize=axis_sz-3, labelpad = ylabpad)
ax.set_xlabel(xLab, fontsize=axis_sz-3, labelpad = xlabpad)
# reverse axis of y is depth
if not _basemap:
ax = make_section(plt.gca(), None, _x,_y, _latLon_params, 'raw')
cbar_pad = .22
# create colorbar
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3%", pad=cbar_pad)
cbar = plt.colorbar(CS, cax=cax)
cbar_label = _in_var_names[0] + ' (' + units_dict[_in_var_names[0]] + ')'
cbar.ax.set_ylabel(cbar_label, fontsize=axis_sz)
# set tick size
xtickNames = ax.get_xticklabels()
ytickNames = ax.get_yticklabels()
cbartickNames = cbar.ax.get_yticklabels()
for names in [ytickNames, xtickNames, cbartickNames]:
plt.setp(names, rotation=0, fontsize=tick_sz-4)
# plt.show()
return fig
